
MSEI
Molecular structure elucidation by integrating different data mining strategies

Abstract
The overall goal of this project is to develop and implement advanced data-driven programming tools, enabling a superior insight into ultra-high performance liquid chromatography coupled to high- resolution mass spectrometry (UHPLC-HRMS) data. While HPLC has been used as the first level of analyte separation since the 1960s, HRMS is a relatively new and powerful analytic technique used for discovery of molecular species based on their exact mass to charge ratio (m/z). The instrumentation applied is capable of separating mass fragments at the fourth or fifth decimal place. The additional information narrows down the possible chemical formulas of a molecule and thus allows an unprecedented unambiguous qualitative and quantitative assessment of the composition of various types of samples. Not surprisingly, HRMS has found applications across a broad spectrum of scientific fields.
Although we can routinely discern hundreds to thousands of molecular ‘features’ in complex samples such as blood, aerosols, soil, or biofuels, the complexity of the resulting data stream increases proportionally, producing millions of data points per second in multidimensional space. Thus post-processing and data reduction methods followed by data mining and innovative visualization techniques are required to yield meaningful information from HRMS. The project is about developing semi-automatic methods to confidently pinpoint each unknown molecular structure. It is a unique opportunity to expand the applicability of both HRMS and the Kendrick Mass Defect (KMD) approach beyond their current state-of-the-art applications, as well as beyond the capabilities of other analytic methods such as NMR and X-ray crystallography tools that typically require pure samples in relatively large amounts.
People
Collaborators


Eliza has joined the academic team as a senior scientist. She previously worked as a postdoctoral researcher at the Massachusetts Institute of Technology (2012-2013), Empa (2013-2017) and the University of Innsbruck (2017-2020). Eliza received her PhD in Atmospheric Science from the Max Planck Institute for Chemistry in 2012, and her Bachelor degree with Honours in Antarctic Science from the University of Tasmania in 2008. Her previous research has centered around the use of novel isotopic measurements and modelling approaches in atmospheric and biogeosciences, in particular the nitrogen cycle. Her research at SDSC will focus on data analytics and machine learning approaches in environmental and natural sciences.


Fernando Perez-Cruz received a PhD. in Electrical Engineering from the Technical University of Madrid. He is Titular Professor in the Computer Science Department at ETH Zurich and Head of Machine Learning Research and AI at Spiden. He has been a member of the technical staff at Bell Labs and a Machine Learning Research Scientist at Amazon. Fernando has been a visiting professor at Princeton University under a Marie Curie Fellowship and an associate professor at University Carlos III in Madrid. He held positions at the Gatsby Unit (London), Max Planck Institute for Biological Cybernetics (Tuebingen), and BioWulf Technologies (New York). Fernando Perez-Cruz has served as Chief Data Scientist at the SDSC from 2018 to 2023, and Deputy Executive Director of the SDSC from 2022 to 2023


Guillaume Obozinski graduated with a PhD in Statistics from UC Berkeley in 2009. He did his postdoc and held until 2012 a researcher position in the Willow and Sierra teams at INRIA and Ecole Normale Supérieure in Paris. He was then Research Faculty at Ecole des Ponts ParisTech until 2018. Guillaume has broad interests in statistics and machine learning and worked over time on sparse modeling, optimization for large scale learning, graphical models, relational learning and semantic embeddings, with applications in various domains from computational biology to computer vision.


Lili obtained the MSc in Statistics from ETH in 2018. She wrote her Master thesis at the Swiss Data Science Center applying topic modelling to political data. She rejoined the center in May 2020 after a year as a statistical consultant at the Seminar for Statistics at ETH. With her MSc in Chemical Engineering, she worked as a process engineer in the glass industry for several years. She is interested in interdisciplinary projects where data science can help uncover new insights.


Michele received a Ph.D. in Environmental Sciences from the University of Lausanne (Switzerland) in 2013. He was then a visiting postdoc in the CALVIN group, Institute of Perception, Action and Behaviour of the School of Informatics at the University of Edinburgh, Scotland (2014-2016). He then joined the Multimodal Remote Sensing and the Geocomputation groups at the Geography department of the University of Zurich, Switzerland (2016-2017). His main research activities were at the interface of computer vision, machine and deep learning for the extraction of information from aerial photos, satellite optical images and geospatial data in general.
description
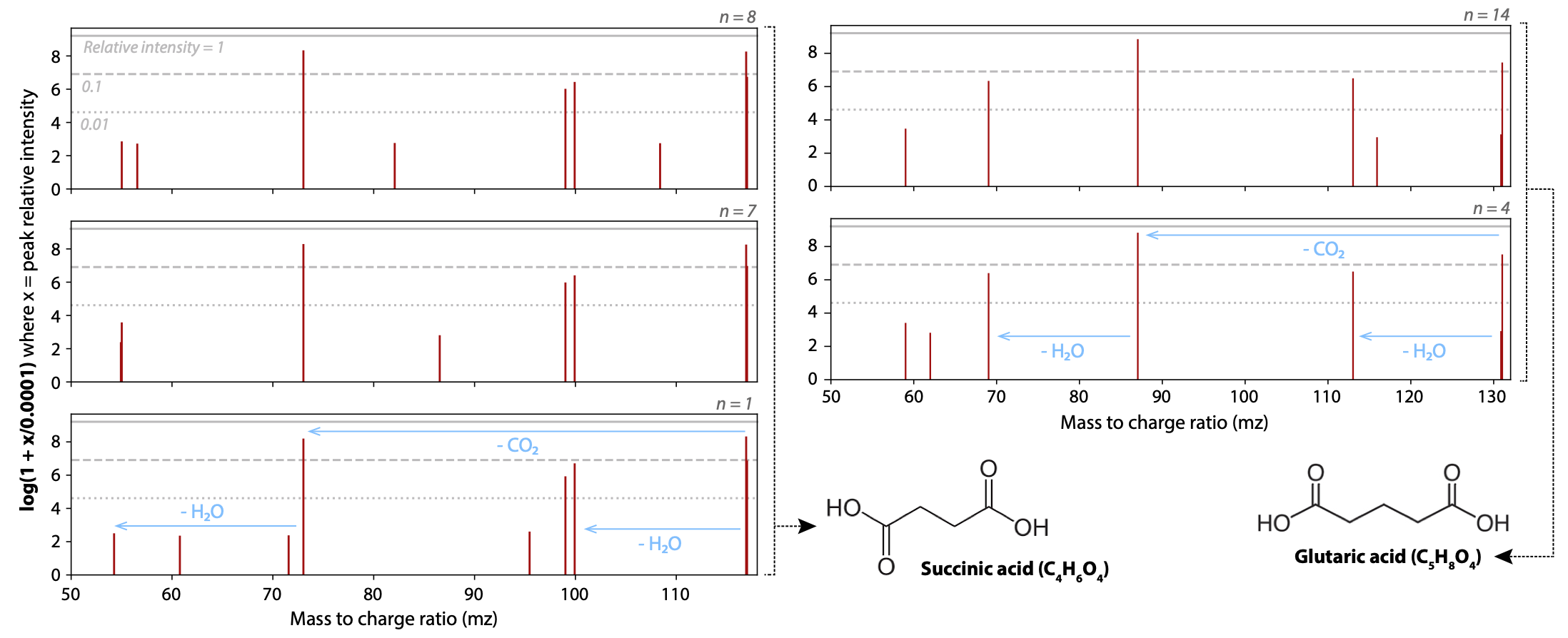
- Molecular clustering based on UHPLC-HRMS/MS data reflecting chemical “families” based on the presence of similar functional groups.
- Within-cluster prediction of functional groups and molecular structure for unknown compounds.
- Predictive modelling of molecular fragmentation patterns, retention time, and other features.
Presentation
Gallery
Annexe
Additionnal resources
Bibliography
- Wu et al. (2021) Valence Photoionization and Energetics of Vanillin, a Sustainable Feedstock Candidate, The Journal of Physical Chemistry A, doi: 10.1021/acs.jpca.1c00876
- Dührkop et al. (2020) Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra, Nature Biotechnology, doi: 10.1038/s41587-020-0740-8
- Arturi et al. (2019) Molecular footprint of co-solvents in hydrothermal liquefaction (HTL) of Fallopia Japonica, Journal of Supercritical Fluids, doi: 10.1016/j.supflu.2018.08.010
- Roach et al. (2011) Higher-Order Mass Defect Analysis for Mass Spectra of Complex Organic Mixtures, Analytical Chemistry, doi: 10.1021/ac200654j
Publications
Related Pages
More projects
CLIMIS4AVAL
News
Latest news

Efficient and scalable graph generation through iterative local expansion
Efficient and scalable graph generation through iterative local expansion


RAvaFcast | Automating regional avalanche danger prediction in Switzerland
RAvaFcast | Automating regional avalanche danger prediction in Switzerland


PassGPT | Using language models to enhance password security
PassGPT | Using language models to enhance password security
Contact us
Let’s talk Data Science
Do you need our services or expertise?
Contact us for your next Data Science project!



